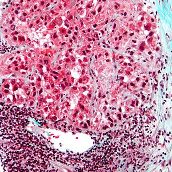
Hepatocellular carcinoma genomic data from the Asia Cancer Research Group.
Dataset type: Genomic
Data released on May 31, 2012
Kan Z; Zheng H; Liu X; Li S; Barber TD; Gong Z; Gao H; Hao K; Willard MD; Xu J; Hauptschein R; Rejto PA; Fernandez J; Wang G; Zhang Q; Wang B; Chen R; Wang J; Lee NP; Lee WH; Ariyaratne PN; Tennakoon C; Mulawadi FH; Wong KF; Liu AM; Chan KL; Hu Y; Chou W; Buser C; Zhou W; Lin Z; Peng Z; Yi K; Chen S; Li L; Fan X; Yang J; Ye R; Ju J; Wang K; Estrella H; Deng S; Wulur IH; Liu J; Ehsani ME; Zhang C; Loboda A; Sung W; Aggarwal A; Poon RT; Fan ST; Wang J; Hardwick J; Reinhard C; Dai H; Li Y; Luk JM; Mao M; the Asian Cancer Research Group (2012): Hepatocellular carcinoma genomic data from the Asia Cancer Research Group. GigaScience. https://doi.org/10.5524/100034
Hepatocellular carcinoma (HCC) is one of the most common solid tumors worldwide and represents the third leading cause of cancer deaths. Hepatitis B virus (HBV) is a major etiologic agent, leading to an increased risk of developing HCC, in particular those with acute liver disease and cirrhosis. The Asian Cancer Research Group (ACRG) is an independent, not-for-profit company established to accelerate research and improve treatment for patients affected with the most commonly-diagnosed cancers in Asia. With HBV being endemic in China and Southeast Asia, and high levels of HCC being a result, the ACRG have studied the events and effects of HBV integration in the HCC genome. To do this massively parallel sequencing in a cohort of 88 Chinese patients diagnosed with HCC who underwent curative primary hepatectomy or liver transplantation at Queen Mary Hospital (Pokfulam, Hong Kong) was carried out. All patients gave written informed consent to use both tumor (T) and non-tumor (N) liver tissues for the study. Genomic DNA was purified for at least 30-fold coverage paired-end (PE) sequencing, and PE reads were mapped on human reference genome (UCSC build hg19) and HBV (NC_003977). Two sequencing libraries with different insert size were constructed for each genomic DNA sample (200 bp and 800 bp). Paired end, 90bp read length sequencing was performed in the HiSeq 2000 sequencer according to Manufacturer’s instructions. Raw gene expression profiling data of these human HCC samples has also deposited to GEO with the accession number GSE25097.
Additional details
Read the peer-reviewed publication(s):
- Sung, W.-K., Zheng, H., Li, S., Chen, R., Liu, X., Li, Y., Lee, N. P., Lee, W. H., Ariyaratne, P. N., Tennakoon, C., Mulawadi, F. H., Wong, K. F., Liu, A. M., Poon, R. T., Fan, S. T., Chan, K. L., Gong, Z., Hu, Y., Lin, Z., … Luk, J. M. (2012). Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nature Genetics, 44(7), 765–769. https://doi.org/10.1038/ng.2295 (PubMed:22634754)
Accessions (data included in GigaDB):
ERA: ERP001196
Accessions (data not in GigaDB):
GEO: GSE25097
Click on a table column to sort the results.
Table Settings| Sample ID | Common Name | Scientific Name | Sample Attributes | Taxonomic ID | Genbank Name |
|---|---|---|---|---|---|
| 101N | Human | Homo sapiens | Disease status:normal |
9606 | human |
| 101T | Human | Homo sapiens | Disease status:DOID:684 |
9606 | human |
| 105N | Human | Homo sapiens | Disease status:normal |
9606 | human |
| 105T | Human | Homo sapiens | Disease status:DOID:684 |
9606 | human |
| 106N | Human | Homo sapiens | Disease status:normal |
9606 | human |
| 106T | Human | Homo sapiens | Disease status:DOID:684 |
9606 | human |
| 108N | Human | Homo sapiens | Disease status:normal |
9606 | human |
| 108T | Human | Homo sapiens | Disease status:DOID:684 |
9606 | human |
| 11N | Human | Homo sapiens | Disease status:normal |
9606 | human |
| 11T | Human | Homo sapiens | Disease status:DOID:684 |
9606 | human |
Click on a table column to sort the results.
Table Settings| File Name | Description | Sample ID | Data Type | File Format | Size | Release Date | File Attributes | Download |
|---|---|---|---|---|---|---|---|---|
| Genome sequence | FASTQ | 3.51 GB | 2012-05-31 | |||||
| Genome sequence | FASTQ | 3.61 GB | 2012-05-31 | |||||
| Genome sequence | FASTQ | 4.26 GB | 2012-05-31 | |||||
| Genome sequence | FASTQ | 4.23 GB | 2012-05-31 | |||||
| Genome sequence | FASTQ | 4.40 GB | 2012-05-31 | |||||
| Genome sequence | FASTQ | 4.53 GB | 2012-05-31 | |||||
| Genome sequence | FASTQ | 5.30 GB | 2012-05-31 | |||||
| Genome sequence | FASTQ | 5.37 GB | 2012-05-31 | |||||
| Genome sequence | FASTQ | 5.40 GB | 2012-05-31 | |||||
| Genome sequence | FASTQ | 5.41 GB | 2012-05-31 |
| Date | Action |
|---|---|
| October 15, 2015 | File 100914_I137_FC80139ABXX_L1_HUMohgRAADMAAPE_1.clean.fq.gz updated |
| October 15, 2015 | File 100914_I137_FC80139ABXX_L1_HUMohgRAADMAAPE_1.clean.fq.gz updated |
| October 26, 2015 | File 100914_I137_FC80139ABXX_L1_HUMohgRAADMAAPE_1.clean.fq.gz updated |
| November 4, 2015 | File 100914_I137_FC80139ABXX_L1_HUMohgRAADMAAPE_1.clean.fq.gz updated |
| November 4, 2015 | File 100914_I137_FC80139ABXX_L1_HUMohgRAADMAAPE_2.clean.fq.gz updated |
| November 4, 2015 | File 100914_I137_FC80139ABXX_L2_HUMohgRAGDMAAPE_1.clean.fq.gz updated |
| November 4, 2015 | File 100914_I137_FC80139ABXX_L2_HUMohgRAGDMAAPE_2.clean.fq.gz updated |
| November 4, 2015 | File 100914_I137_FC80139ABXX_L3_HUMohgRAIDMAAPE_1.clean.fq.gz updated |
| November 4, 2015 | File 100914_I137_FC80139ABXX_L3_HUMohgRAIDMAAPE_2.clean.fq.gz updated |
| November 4, 2015 | File 100914_I137_FC80139ABXX_L4_HUMohgRANDMAAPE_1.clean.fq.gz updated |
| November 4, 2015 | File 100914_I137_FC80139ABXX_L4_HUMohgRANDMAAPE_2.clean.fq.gz updated |
| November 4, 2015 | File 100914_I137_FC80139ABXX_L5_HUMohgRBDDMAAPE_1.clean.fq.gz updated |
| November 4, 2015 | File 100914_I137_FC80139ABXX_L5_HUMohgRBDDMAAPE_2.clean.fq.gz updated |
| November 4, 2015 | File 100914_I137_FC80139ABXX_L5_HUMohgRBDDMAAPE_2.clean.fq.gz updated |
| November 4, 2015 | File 101222_I327_FC810M9ABXX_L8_HUMohgRHLDMAAPEI-8_2.clean.fq.gz updated |
| March 2, 2016 | Link updated : ERA:ERP001196 |